Page Summary
-
This unit uses the Palmer Penguins dataset and the YDF library to train and interpret a decision tree for species prediction.
-
Decision trees support numerical, categorical, and missing features without preprocessing, offering inherent interpretability.
-
Hyperparameter tuning, including adjusting
min_examples, can significantly improve decision tree performance. -
While often less accurate than complex models, decision trees serve as valuable baselines and tools for interpretability.
-
Random forests, discussed in subsequent sections, generally provide superior predictive accuracy compared to single decision trees.
In this unit, you'll use the YDF (Yggdrasil Decision Forest) library train and interpret a decision tree.
This unit is inspired from the 🧭 YDF Getting Started tutorial.
Preliminaries
Before studying the dataset, do the following:
- Create a new Colab notebook.
- Install the YDF library by placing and executing the following line of code
in your new Colab notebook:
!pip install ydf -U - Import the following libraries:
import ydf import numpy as np import pandas as pd
The Palmer Penguins dataset
This Colab uses the Palmer Penguins dataset, which contains size measurements for three penguin species:
- Chinstrap
- Gentoo
- Adelie
This is a classification problem—the goal is to predict the species of penguin based on data in the Palmer's Penguins dataset. Here are the penguins:

Figure 16. Three different penguin species. Image by @allisonhorst
The following code calls a pandas function to load the Palmer Penguins dataset into memory:
path = "https://storage.googleapis.com/download.tensorflow.org/data/palmer_penguins/penguins.csv"
dataset = pd.read_csv(path)
label = "species"
# Display the first 3 examples.
dataset.head(3)
The following table formats the first 3 examples in the Palmer Penguins dataset:
Table 3. The first 3 examples in Palmer Penguins
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | male | 2007 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | female | 2007 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | female | 2007 |
The full dataset contains a mix of numerical (for example, bill_depth_mm),
categorical (for example, island), and missing features. Unlike neural
networks, decision forests support all these feature types natively, so you
don't have to do one-hot encoding, normalization, or extra is_present feature.
The following code cell splits the dataset into a training set and testing set:
# Use the ~20% of the examples as the testing set
# and the remaining ~80% of the examples as the training set.
np.random.seed(1)
is_test = np.random.rand(len(dataset)) < 0.2
train_dataset = dataset[~is_test]
test_dataset = dataset[is_test]
print("Training examples: ", len(train_dataset))
# >> Training examples: 272
print("Testing examples: ", len(test_dataset))
# >> Testing examples: 72
Training a decision trees with default hyperparameters
You can train your first decision tree with the CART (Classification and
Regression Trees) learning algorithm (a.k.a. learner) without specifying any
hyperparameters.
That's because the ydf.CartLearner learner provides good default
hyperparameter values. You will learn more about how this type of model works
later in the course.
model = ydf.CartLearner(label=label).train(train_dataset)
The preceding call did not specify columns to use as input features. Therefore, every column in the training set is used. The call also did not specify the semantics (for example, numerical, categorical, text) of the input features. Therefore, feature semantics are automatically infer.
Call model.plot_tree() to display the resulting decision tree:
model.plot_tree()
In Colab, you can use the mouse to display details about specific elements such as the class distribution in each node.
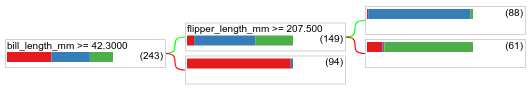
Figure 17. A decision tree trained with default hyperparameters.
Colab shows that the root condition contains 243 examples. However, you might remember that the training dataset contained 272 examples. The remaining 29 examples have been reserved automatically for validation and the tree pruning.
The first condition tests the value of bill_depth_mm. Tables 4 and 5 show the
likelihood of different species depending on the outcome of the first condition.
Table 4. Likelihood of different species if bill_depth_mm ≥
42.3
| Species | Likelihood |
|---|---|
| Adelie (red) | 8% |
| Gentoo (blue) | 58% |
| Chinstrap (green) | 36% |
Table 5. Likelihood of different species if
bill_depth_mm < 42.3
| Species | Likelihood |
|---|---|
| Adelie (red) | 97% |
| Gentoo (blue) | 2% |
| Chinstrap (green) | 0% |
bill_depth_mm is a numerical feature. Therefore, the value 42.3 was found
using the exact splitting for binary classification with numerical
features algorithm.
If bill_depth_mm ≥ 42.3 is True, further testing whether the
flipper_length_mm ≥ 207.5 can almost perfectly separate the
Gentoos and from the Gentoos+Adelie.
The following code provides the training and test accuracy of the model:
train_evaluation = model.evaluate(train_dataset)
print("train accuracy:", train_evaluation.accuracy)
# >> train accuracy: 0.9338
test_evaluation = model.evaluate(test_dataset)
print("test accuracy:", test_evaluation.accuracy)
# >> test accuracy: 0.9167
It is rare, but possible, that the test accuracy is higher than the training accuracy. In that case, the test set possibly differs from the training set. However, this is not the case here as the train & test was split randomly. A more likely explanation is that the test dataset is very small (only 72 examples), so the accuracy estimation is noisy.
In practice, for such a small dataset, using cross-validation would be preferable as it would compute more accurate evaluation metric values. However, in this example, we continue with a training & testing for the sake of simplicity.
Improving the model hyperparameters
The model is a single decision tree was trained with default hyperparameter values. To obtain better predictions, you can:
Use a more powerful learner such as a random forest or a gradient boosted trees model. Those learning algorithms will be explained in the next page.
Optimizer the hyperparameter using your observations and intuiting. The model improvement guide can be helpful.
Use hyperparameter tuning to test automatically a large number of possible hyperparameters.
Since we have not yet seen the random forest and gradient boosted trees algorithm, and since the number of examples is too small to do an automatic hyperparameter tuning, you will manually improve the model.
The decision tree shown above is small, and the leaf with 61 example contains a mix of Adelie and Chinstrap labels. Why didn't the algorithm divide this leaf further? There are two possible reasons:
- The minimum number of samples per leaf (
min_examples=5by default) may have been reached. - The tree might have been divided and then pruned to prevent overfitting.
Reduce the minimum number of examples to 1 and see the results:
model = ydf.CartLearner(label=label, min_examples=1).train(train_dataset)
model.plot_tree()
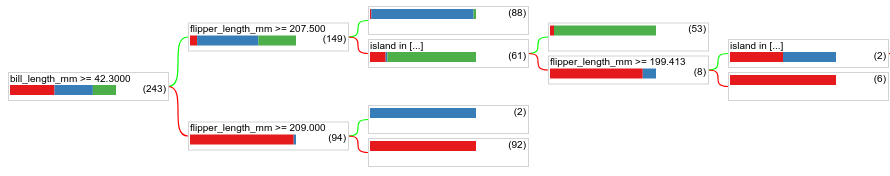
Figure 18. A decision tree trained with min_examples=1.
The leaf node containing 61 examples has been further divided multiple times.
To see if further dividing the node is valuable, we evaluate the quality of this new model on the test dataset:
print(model.evaluate(test_dataset).accuracy)
# >> 0.97222
The quality of the model has increased with a test accuracy going from 0.9167 to 0.97222. This change of hyperparameter was a good idea.
Preview of decision forests
By continuing to improve the hyperparameters, we could likely reach a perfect accuracy. However, instead of this manual process, we can train a more powerful model such as a random forest and see if it works better.
model = ydf.RandomForestLearner(label=label).train(train_dataset)
print("Test accuracy: ", model.evaluate(test_dataset).accuracy)
# >> Test accuracy: 0.986111
The accuracy of the random forest is better than our simple tree. You will learn in the next pages why.
Usage and limitation
As mentioned earlier, a single decision tree often has lower quality than modern machine learning methods like random forests, gradient boosted trees, and neural networks. However, decision trees are still useful in the following cases:
- As a simple and inexpensive baseline to evaluate more complex approaches.
- When there is a tradeoff between the model quality and interpretability.
- As a proxy for the interpretation of the decision forests model, which the course will explore later on.