Sandbox for playing with Sparse JAX and the sparse compiler on CPU.
See colab. Currently, it is only available to googlers.
In particular, this runs the colab [jaxpruner] Sparse Model ViT, written by Utku Evci.
Setup
import functools
import jax
# This is required to get Sparse JAX (experimental for now).
from jax.experimental import sparse
import jax.numpy as jnp
import os
from colabtools import adhoc_import
CITC_CLIENT = ""
CITC_USER = ""
CL_NUMBER = "HEAD"
# Use XLA-NEXT for fast sparse ops
# --xla_cpu_use_xla_runtime enables XLA-NEXT which has fast sparse ops
# --xla_cpu_enable_mlir_tiling_and_fusion=false needed to workaround a few bugs.
# Eventually, it should be removed
os.environ['XLA_FLAGS'] = '--xla_cpu_use_xla_runtime --xla_cpu_enable_mlir_tiling_and_fusion=false'
Importing modules from P4 HEAD.
JAXPRUNER Imports
import tensorflow as tf
import flax
import time
import matplotlib.pyplot as plt
import pandas as pd
JAXPRUNER: TF version = 2.15.0
JAXPRUNER Helper Methods
PRUNE_THRESHOLD = 0.06
def initialize_model(model, placeholder_input, rngs):
"""Initialization function to be jitted."""
init_model_state, init_params = model.flax_model.init(
rngs, placeholder_input, train=False, debug=False
).pop('params')
return init_params, init_model_state
def filtered_bcoo_simple(key, param):
if key[-1] == 'kernel' and 4 > param.ndim > 1:
# Aart made this pruning up, this has no ML semantics AFAIK.
# param = jnp.where(abs(param) < PRUNE_THRESHOLD, jnp.zeros(param.shape), param)
return sparse.BCOO.fromdense(param)
else:
return param
def total(val):
return functools.reduce((lambda x, y: x * y), val)
JAXPRUNER Model Initialization
config = imagenet_vit_config.get_config()
dataset_meta_data = {
'input_dtype': jax.numpy.float32,
'input_shape': (-1, 224, 224, 3),
'num_classes': 1000,
'num_eval_examples': 50000,
'num_train_examples': 1281167,
'target_is_onehot': False,
}
model_cls = models.get_model_cls(config.model_name)
model = model_cls(config, dataset_meta_data)
rng, init_rng = jax.random.split(jax.random.PRNGKey(8))
# Initialize model.
rng, init_rng = jax.random.split(rng)
placeholder_input = jnp.ones((1, 224, 224, 3))
init_params, init_model_state = initialize_model(
model, placeholder_input, {'params': init_rng}
)
initial_train_state = train_utils.TrainState(
global_step=0, params=init_params, model_state=init_model_state, rng=rng
)
# Uses hardcoded cns for testing.
init_checkpoint_path = (
'/cns/vz-d/home/brain-sparsity/evcu/sparsevit-checkpoints/sparse_b16_80/'
)
restored_train_state = pretrain_utils.restore_pretrained_checkpoint(
init_checkpoint_path, initial_train_state, assert_exist=True
)
res_dict = {}
for k, p in flax.traverse_util.flatten_dict(restored_train_state.params).items():
res_dict[k] = filtered_bcoo_simple(k, p)
sparse_params = flax.traverse_util.unflatten_dict(res_dict)
i = 0
for k, v in flax.traverse_util.flatten_dict(sparse_params).items():
if isinstance(v, sparse.BCOO):
density = v.nse / total(v.shape)
print('JAXPRUNER: BCOO @', i, k[-2:], v.shape, v.nse, density)
i = i + 1
# Get the variables.
variables = {'params': restored_train_state.params, **init_model_state}
sp_variables = {'params': sparse_params, **init_model_state}
print('JAXPRUNER: ready to run from', init_checkpoint_path)
JAXPRUNER Dense Run
start_time = time.time()
lowered = jax.jit(
functools.partial(model.flax_model.apply, train=False)
).lower(variables, jnp.ones((1, 224, 224, 3)))
compiled = lowered.compile()
dense_c_time = time.time() - start_time
print('JAXPRUNER: dense compiled %s seconds ---' % (dense_c_time))
start_time = time.time()
res_dense = compiled(variables, jnp.ones((1, 224, 224, 3)))
dense_r_time = time.time() - start_time
print('JAXPRUNER: dense run %s seconds ---' % (dense_r_time))
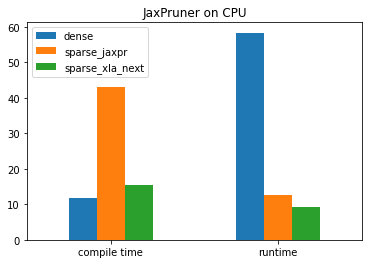
JAXPRUNER: dense compiled 11.863374471664429 seconds --- JAXPRUNER: dense run 58.369417905807495 seconds ---
JAXPRUNER Sparse Run (using pure JAXPR)
start_time = time.time()
sparse_lower = (
jax.jit(
sparse.sparsify(
functools.partial(model.flax_model.apply, train=False)
)
)
.lower(sp_variables, jnp.ones((1, 224, 224, 3)))
)
sparse_apply = sparse_lower.compile()
sparse_jaxpr_c_time = time.time() - start_time
print('JAXPRUNER: sparse JAXPR compiled %s seconds ---' % (sparse_jaxpr_c_time))
start_time = time.time()
res_jaxpr = sparse_apply(sp_variables, jnp.ones((1, 224, 224, 3)))
sparse_jaxpr_r_time = time.time() - start_time
print('JAXPRUNER: sparse JAXPR run %s seconds ---' % (sparse_jaxpr_r_time))
JAXPRUNER: sparse JAXPR compiled 43.14039134979248 seconds --- JAXPRUNER: sparse JAXPR run 12.49958348274231 seconds ---
JAXPRUNER Sparse Run (using XLA-Next Sparse Compiler)
start_time = time.time()
sparse_lower = (
sparse_jit(functools.partial(model.flax_model.apply, train=False)
)
.lower(sp_variables, jnp.ones((1, 224, 224, 3)))
)
sparse_apply = sparse_lower.compile()
sparse_xla_next_c_time = time.time() - start_time
print('JAXPRUNER: sparse XLA-NEXT compiled %s seconds ---' % (sparse_xla_next_c_time))
start_time = time.time()
res_xla_next = sparse_apply(sp_variables, jnp.ones((1, 224, 224, 3)))
sparse_xla_next_r_time = time.time() - start_time
print('JAXPRUNER: sparse XLA-NEXT run %s seconds ---' % (sparse_xla_next_r_time))
JAXPRUNER: sparse XLA-NEXT compiled 15.409374713897705 seconds --- JAXPRUNER: sparse XLA-NEXT run 9.1929030418396 seconds ---
JAXPRUNER Results
axis = ['dense', 'sparse_jaxpr', 'sparse_xla_next']
data1 = [dense_c_time, sparse_jaxpr_c_time, sparse_xla_next_c_time]
data2 = [dense_r_time, sparse_jaxpr_r_time, sparse_xla_next_r_time]
df = pd.DataFrame([data1, data2], index=['compile time','runtime'], columns=axis)
df.plot.bar(rot=0).set_title("JaxPruner on CPU")
Text(0.5, 1.0, 'JaxPruner on CPU')